2026-01-27
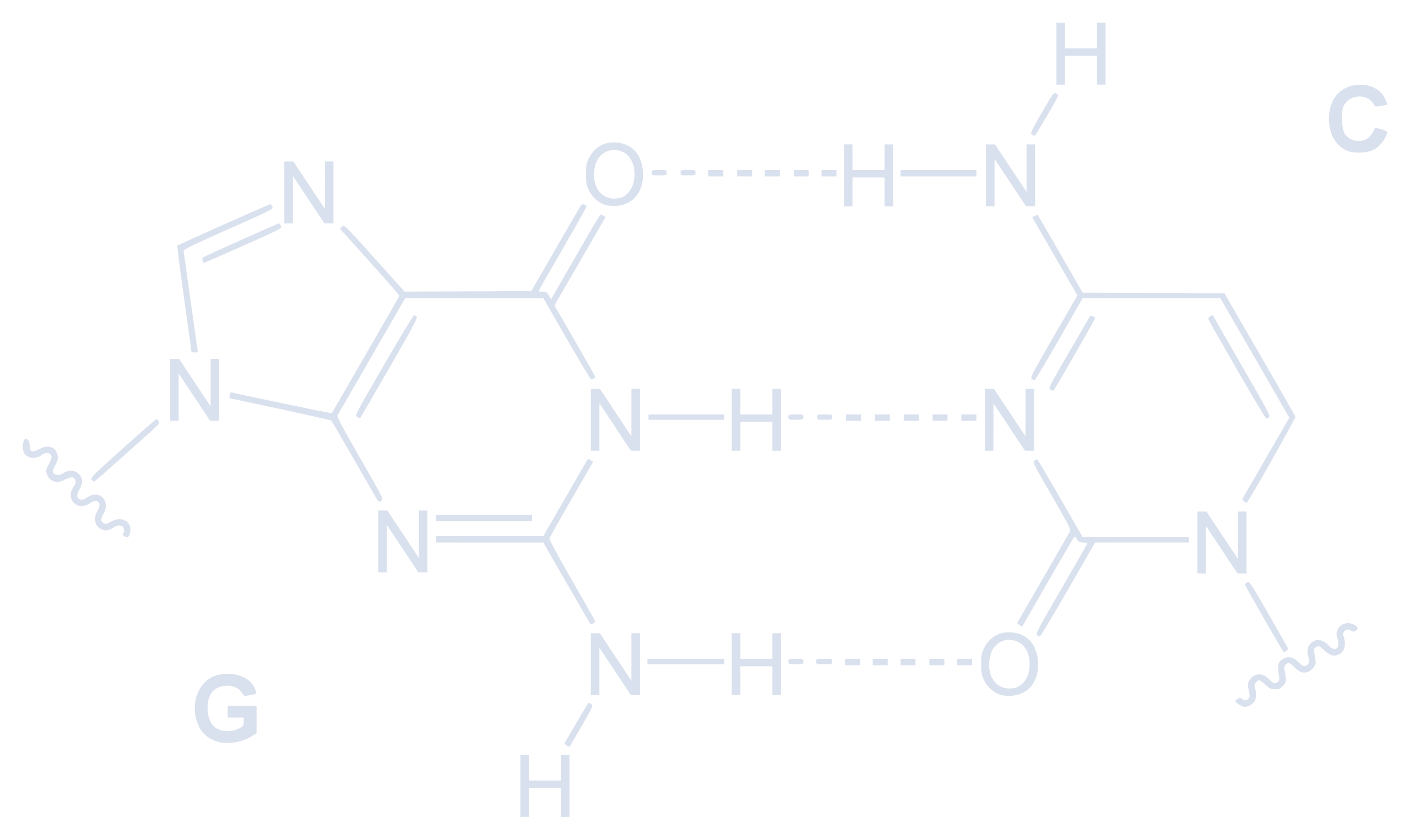
Our review on Machine Learning for #RNA Secondary Structure Prediction is now published in RNA Society Journal ! dx.doi.org/10.1261/rna.... Summary in this previous thread bsky.app/profile/buss...
(view post)2025-12-24
New #preprint alert! 🧬 MERGE-RNA: a physics-based model to predict #RNA secondary structure ensembles with chemical probing, lead by Giuseppe Sacco, in collaboration with Guido Sanguinetti and Redmond Smyth's lab arxiv.org/abs/2512.20581
(view post)2025-12-10
Our work presented today at the XBW2025 workshop homepage.uni-graz.at/de/georg.pab...! Maurizio Gilioli presented our recent #MD simulations of nucleoproteins from influenza A virus
(view post)2025-11-28
🎉Our group has been awarded a prestigious #FIS grant! #RNAScale - Exploring RNA Dynamics across Scales: From Base Pairs to Viral Genomes 👉 www.bussilab.org/fis3
We will soon open postdoctoral positions across several components of the project! An expression-of-interest form is already online.
(view post)2025-11-08
The work describing the #CASP16 solvation challenge has been published in Proteins journal! doi.org/10.1002/prot...
(view post)2025-11-06
Our work presented at the Ultra-Highfield NMR and 4D Structural Biology workshop www.bnmrz.org/4dstructbiol organized by Sattler Lab! Giovanni Bussi will show our works on #RNA dynamics integrating #MD, #SAXS, and #NMR and validating a #CASP16 target using #cryoEM data
(view post)2025-11-05
New #preprint alert! 🧬 Machine Learning for #RNA Secondary Structure Prediction: a #review of current methods and challenges, by Giuseppe Sacco, Giovanni Bussi, and Guido Sanguinetti arxiv.org/abs/2511.02622
(view post)2025-11-05
A new tutorial manuscript is out! 🚀 Making PLUMED fly: a tutorial on optimizing performance, by Daniele Rapetti, Max Bonomi Carlo Camilloni Giovanni Bussi and Gareth Tribello. arxiv.org/abs/2511.02699
(view post)2025-11-04
A recent manuscript describing our and others’ efforts to predict the solvation shell of a group I intron is now available on bioRxiv! 🔬 doi.org/10.1101/2025...
(view post)2025-11-04
📽️ The recording of Giovanni Bussi 's talk at CASP RNA Special Interest Group is now available at www.youtube.com/watch?v=NRCg...
(view post)2025-10-30
Our work presented today at the www.youtube.com/CASPRNASIG online seminar! Giovanni Bussi will report on the #CASP16 solvation challenge and 2D ensemble reconstructions using #DMS data
(view post)2025-10-13
Our work presented today at the Biophysical Modeling of the Structure and Function of Viruses CECAM workshop at the Institut d'Etudes Scientifiques de Cargèse! Giovanni Bussi will report on the recent #CASP16 solvation challenge and 2D ensemble reconstructions using #DMS data
(view post)2025-09-26
Congratulations to Elisa Posani for successfully defending her PhD thesis "Ensemble Refinement of Cryo-Electron Microscopy-Derived RNA Structures Using Molecular Dynamics Simulations"! Cosupervision with Alessandra Magistrato
(view post)2025-09-26
Congratulations to Ivan Gilardoni for successfully defending his PhD thesis "Development and application of methods to integrate molecular simulations with experimental measurements"!
(view post)2025-09-19
Congratulations to Davide Di Vora for successfully defending his master thesis on the "On-the-fly integration of experimental data and molecular simulations" at University of Trieste! 🥳
(view post)2025-09-10
Our work presented today at the ICTP Conference on Frontiers in Atomistic Simulations: from Physics to Chemistry and Biology! Olivier Languin-Cattoën and Ivan Gilardoni will share posters with our results on enhancing #RNA - Mg binding and on integrating #MD simulations and experiments
(view post)2025-08-21
Our research presented at the Cargese school on Molecular Evolution and Design! Salvo Di Marco will show our simulations of #RNA #membrane interactions and Elisa Posani will share our approach integrating #MD and #cryoEM to investigate the #dynamics of a group I intron
(view post)2025-08-02
New #openreview from our group at prereview.org/reviews/1672...! Salvo Di Marco and Giovanni Bussi reviewed a #preprint by Davit Potoyan on the Molecular Drivers of #RNA Phase Separation. Also available at doi.org/10.5281/zeno...
(view post)2025-07-22
Our research presented today at the Telluride meeting on #RNA dynamics! Giovanni Bussi will report on some of our ongoing projects: the recent #CASP16 solvation challenge, constant pH #MD simulations on #RNA, and 2D ensemble reconstructions using #DMS data
(view post)2025-07-22
A review reporting on the discussions held at the recent CECAM workshop on enzymatic plastic degradation that we co-organized in Trieste is now available on #arxiv
(view post)2025-07-16
Our research presented at the Cambridge Ellis Unit Summer School on Probabilistic Machine Learning! Giuseppe Sacco will present a poster reporting our machine-learned model for DMS-guided ensemble prediction of RNA systems www.ellis.eng.cam.ac.uk/summer-schoo...
(view post)2025-07-14
📢 New #preprint alert! Our review "RNA Dynamics and Interactions Revealed through Atomistic Simulations" is now available on arxiv.org/abs/2507.08474 ! Written by Giovanni Bussi and Olivier Languin-Cattoën
(view post)2025-07-14
Our research presented at #StatPhys29! Ivan Gilardoni will present a poster reporting on our methodological work in integrating molecular simulations and experimental data statphys29.org
(view post)2025-06-26
📢 Paper with Sattler Lab now published in Nucleic Acids Research ! doi.org/10.1093/nar/... We combine #NMR, #SAXS, and #MD to construct conformational ensembles, showing that A-to-I hyper-editing leads to significant structural dynamics. MD simulations by Valerio Piomponi
(view post)2025-06-26
A paper from our group (doi.org/10.1093/narg...) has been tested and reviewed in ReproHack www.reprohack.org/review/151! We got interesting feedback on how to improve documentation of our notebooks. Very useful initiative!
(view post)2025-06-25
New #openreview from our group at prereview.org/reviews/1573...! Giovanni Bussi and Olivier Languin-Cattoën reviewed a #preprint by Simon Olsson showing their Minimum-Excess-Work Guidance method. Also available at doi.org/10.5281/zeno...
(view post)2025-06-23
Our research presented at the proteinelectrostatics.org Protein Electrostatics 2025, Lisbon! Tomas Silva shared our constant pH simulations on #RNA systems, from oligomers to introns!
(view post)2025-06-18
Our research presented at the meeting of the Division of Chemistry of Biological Systems of the Società Chimica Italiana! Elisa Posani will show our integrative modelling of #RNA using #cryoEM data and Salvo Di Marco will share our simulations of #RNA membrane interactions
(view post)2025-06-04
Our research presented at the GRC on computational #NMR organized by Katja Petzold in Les Diablerets! Giovanni Bussi will share our integrative approach to reconstruct the impact of A-to-I modifications on dynamics and our protocol to predict Mg2+ binding sites in #RNA molecules
(view post)2025-05-27
📢 Paper on the RBP Footprint Grand Challenge—launched at RNA2021 — is out in RNA Society Journal doi.org/10.1261/rna....! A community-driven benchmark of methods for predicting RBP binding sites, with contributions from Thorben Froehlking, Mattia Bernetti, and Giovanni Bussi
(view post)2025-05-21
Congratulations to Tomas Silva for winning the best poster prize at the #RNA modelling across scales CECAM workshop, describing our constant pH simulations of RNA systems 👏
(view post)2025-05-21
Our research was featured at the CECAM meeting #RNA modelling across scales at SISSA, Trieste! Olivier Languin-Cattoën gave a talk on our enhanced sampling strategy for exploring RNA-Mg2+ binding, and all other group members presented posters on their research
(view post)2025-05-16
Our integrative approach using #MD and #cryoEM data to construct structural ensembles of #RNA just published on doi.org/10.1038/s414... Lead by Elisa Posani, with Alessandra Magistrato Max Bonomi ꙮ Pavel Janoš ꙮ Navtej Toor and Daniel Haack 🎉
(view post)2025-05-15
The manuscript describing MDRefine, our #python package for integrating experimental data and #moleculardynamics trajectories using reweighting methods, has been published on J Chem Phys doi.org/10.1063/5.02...!
(view post)2025-04-17
📢 Ten days left to apply for a #PhD in Molecular and Statistical Biophysics at SISSA 👇 Deadline is April 28!
(view post)2025-04-09
Molecular simulations showing how #RNA structure is affected by biomolecular condensates now published in Proceedings of the National Academy of Sciences www.pnas.org/doi/10.1073/...! Work lead by Matteo Boccalini under the supervision of Alessandro Barducci and Matteo Paloni
(view post)2025-04-08
📢 Apply for the #PhD 🎓 in Molecular and Statistical Biophysics at SISSA in lovely Trieste! 4 fully funded fellowships starting Oct 2025. ~8 months of classes, then choose your project. Remote entrance exam. Application deadline: 28.4.2025. Please repost! 👉 msb.sissa.it/join-us/ #joinus
(view post)2025-04-05
Our community letter on #FAIR principles in #moleculardynamics simulations has been published on naturemethods.bsky.social 🎉
(view post)2025-04-02
Our research presented at an EMBL-EBI workshop on Computational approaches to predict small-molecule/RNA interaction! Today Giovanni Bussi will show our #CASP16 results on predicting Mg binding sites and our attempts to model chemical probing experiments
(view post)2025-03-18
Constant pH #metadynamics simulation of #RNA oligomers now published on American Chemical Society JCIM! Work done by Tomas Silva with some help from Giovanni Bussi . doi.org/10.1021/acs.jcim.4c02185
(view post)2025-03-09
All-atom simulations of #RNA - membrane interactions accepted on American Chemical Society Nano Letters doi.org/10.1021/acs.... ! Work done by Salvo Di Marco under the supervision of Jana Aupic and Alessandra Magistrato
(view post)2025-03-06
Our Research presented at the Chemistry Department of the Ecole Normale Superieure, Paris. Giovanni Bussi will present our efforts to refine force fields for modified nucleotides, to simulate #RNA at constant pH, and to enhance Mg-RNA sampling #casp16. Host: Guillaume Stirnemann
(view post)2025-03-05
Our research presented today at Institut Pasteur | 130 years of biomedical research ! Giovanni Bussi will give a seminar sharing our approaches to reconstruct #RNA dynamics using #moleculardynamics, #cryoEM, secondary structure prediction, and #DMS probing. Host Max Bonomi
(view post)2025-03-04
Our effort to collect and organize #plumed tutorials has been published in the Journal of Chemical Physics, as part of the Michele Parrinello Festschrift!
(view post)2025-01-22
Our research presented today at AreaSciencePark! Giovanni Bussi will discuss our recent results in enhanced sampling of #RNA Mg interactions, integration of #cryoEM data, and refinement of force fields for modified nucleotides. www.areasciencepark.it/en/scientifi...
(view post)2025-01-16
Our research presented at Freie Universität Berlin ! Giovanni Bussi will give a CRC colloquium presenting our recent applications of integrative methods and enhanced sampling to #RNA dynamics. Host: Cecilia Clementi www.mi.fu-berlin.de/en/sfb1114/e...
(view post)2025-01-15
📢 Post-doc opportunity alert! Join the Molecular and Statistical Biophysics group at SISSA. Choose your mentor from Giovanni Bussi, Cristian Micheletti, or Angelo Rosa, or collaborate across these groups. www.bussilab.org/joinus#postd... #joinus Please reshare!
(view post)2025-01-13
Our research presented at kitkarlsruhe.bsky.social Institute of Physical Chemistry! Today Giovanni Bussi will discuss #RNA, integrative methods, enhanced sampling, and recent PLUMED initiatives www.ipc.kit.edu/english/19_3.... Host Tomas Kubar
(view post)2024-12-19
Our work presented at the 3D-BIOINFO-PT workshop 3d-bioinfo-pt.github.io/workshops/wo... and meeting 3d-bioinfo-pt.github.io/annual_meeti...! Tomas Silva will give a PLUMED tutorial on constant pH #metadynamics and show our latest results on #RNA oligomers (arxiv.org/abs/2410.16064)
(view post)2024-12-18
📢 Presentations from #CASP16 are public. Assessment of the RNA-solvation target by Rachael Kretsch is confirmed: our two submissions ranked 1st and 2nd in CC with #cryo-EM 🥳 Congrats to Olivier Languin-Cattoën, Elisa Posani, Giovanni Bussi www.predictioncenter.org/casp16/doc/p...
(view post)2024-12-18
SISSA SBP alumni workshop is starting today! sites.google.com/view/sbpalum... Many talks from previous members of our group: Francesco Di Palma, Andrea Pérez-Villa, Andrea Cesari, Francesca Cuturello, Nicola Calonaci, and Thorben Froehlking 🎉 🎄⛄
(view post)2024-12-16
The Hitchhiker's Guide to the Conformational Space of #DNA G-#quadruplexes is out on J. Comput. Chem. 🥳 ! Work done in Pavel Banáš group, we contributed to define the enhanced sampling strategy. Read more: doi.org/10.1002/jcc....
(view post)2024-12-16
Our work presented at the XBW2024 workshop people.sissa.it/~michelet/XB...! Salvatore Di Marco will show our results on #RNA - #membrane interactions (see doi.org/10.1101/2024...), and Giovanni Bussi will show our recent #CASP16 experience with RNA and Mg2+
(view post)2024-12-06
📢 New #preprint describing our community effort to share #plumed tutorials arxiv.org/abs/2412.03595 ! Explore the tutorials at www.plumed-tutorials.org
(view post)2024-12-04
Our research presented at the #CASP16 meeting www.predictioncenter.org/casp16! Giovanni Bussi will give a talk showing the results obtained with Olivier Languin-Cattoën and Elisa Posani on the #RNA solvation target.
(view post)2024-12-02
Our research presented at the CECAM workshop www.cecam.org/workshop-det...! Ivan Gilardoni will show a poster about our #python package MDrefine. See also arxiv.org/abs/2411.07798
(view post)2024-11-25
New #openreview from our group at disq.us/p/3145xzw! Giovanni Bussi and Ivan Gilardoni reviewed a #preprint by Paul Robustelli on the determination of conformational ensembles for intrinsically disordered proteins, combining molecular dynamics simulations and experimental data. Also posted at prereview.org/reviews/142...
(view post)2024-11-20
Our first #openreview has been published! disq.us/t/4r5064p Olivier Languin-Cattoën and Giovanni Bussi reviewed a #preprint by Guillaume Stirnemann and Glen Hocky on catch bonding behavior in vinculin-actin unbinding kinetics. Also posted at prereview.org/reviews/142...
(view post)2024-11-16
This summer we used #moleculardynamics simulations and #enhancedsampling to predict the solvation shell of a ribozyme! Results will be announced at the coming #CASP16 conference, to which we will attend to present our approach. Finger crossed 🤞 x.com/BiologyAIDaily/stat...
(view post)2024-11-14
🚀Introducing MDRefine: our new #python package for integrating experimental data with #moleculardynamics trajectories through diverse reweighting methods! Check out the #preprint: arxiv.org/abs/2411.07798. Summary in thread 🧵:
(view post)2024-11-12
🚀Excited to share our #preprint on implementing constant pH metadynamics for #RNA oligomers! Check it out here: arxiv.org/abs/2410.16064. Thread 🧵:
(view post)2024-11-09
📢 Our work on #m6A role in #RNA recognition has been published on @JPhysChem B! doi.org/10.1021/acs.jpcb.... An effort led by @Piompons with help from Miroslav Krepl, @sponer_lab , and @BussiGio Summary at x.com/bussilab/status/178...
(view post)2024-04-24
Excited to share our new #preprint on #m6A's role in #RNA recognition! 🧬 A collaborative effort by @piompons, Miroslav Krepl, @sponer_lab, & @BussiGio. Dive into our findings in this thread 👇
(view post)2024-02-06
And today, Giuseppe Sacco will show how we are analyzing #SHAPE datasets from @DrJ_RNA to build a physical model for the interaction of #RNA molecules with chemical probes. Physics of Biomolecules in Bressanone
(view post)2024-02-05
And Ivan Gilardoni will later discuss our recent work on the combination of force field and ensemble refinement, just published in @JPhysChem pubs.acs.org/doi/10.1021/.... Physics of Biomolecules in Bressanone
(view post)2024-02-05
Our work presented at this amazing workshop in Bressanone! @BussiGio will show today how we combine #MD, denaturation experiments, and #cryoEM data to refine #RNA force fields and ensembles!
(view post)2024-01-26
Our #paper on the combination of force-field and ensembles refinement has been published on @JPhysChem Letters 🥳!! doi.org/10.1021/acs.jpcle... Congratulations Ivan Gilardoni, @TFroehlking, @BussiGio. Summary: x.com/bussilab/status/172...
(view post)2023-11-23
Checkout the #preprint of our latest manuscript on ensemble refinement combined with force-field fitting! arxiv.org/abs/2311.13532 by Ivan Gilardoni,@TFroehlking and @BussiGio. Summary in thread:
(view post)2023-10-26
Congratulations to @Piompons for successfully defending his PhD today @SbpSissa @Sissaschool!! 🍾🍾🍾And many thanks to external examiners Martin Zacharias and Modesto @orozco_lab!
(view post)